Interestingly we found that performing the check sequentially was actually faster than using streams, this post attempts to determine why this is.
Map
|
Sequential Time
|
Device functions
|
Streams
|
32x32
|
1
|
8.341714
|
21.75491
|
64x64
|
3.8
|
16.37582
|
47.9231
|
128x128
|
16.2
|
32.52248
|
133.1749
|
256x256
|
68.01
|
65.31222
|
409.7781
|
512x512
|
277.07
|
140.5209
|
19658.81
|
1024x1024
|
1146.38
|
399.7442
|
144157.2
|
2048x2048
|
4954.72
|
1745.794
|
462554.9
|
Device Functions vs Streams
 |
| Evaluate Neighbours Device Function |
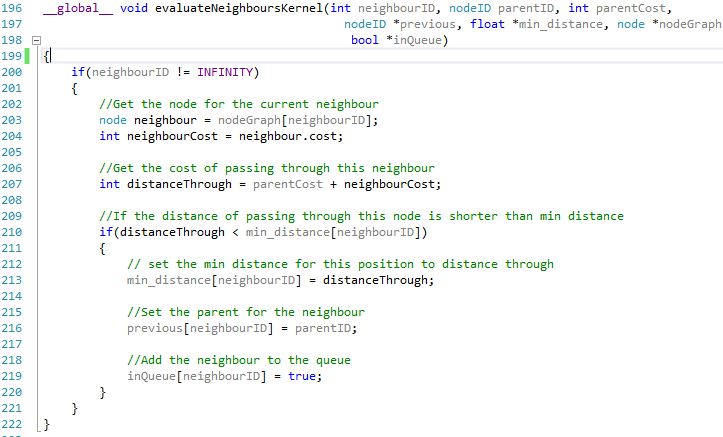 |
| Evaluate Neighbours Kernel |
Looking at the code snippets above they are both almost identical, the only real difference is that one is tagged with the __device__ keyword while the other is tagged with __global__, however this seems to make a big difference.
Map
|
Device Function
|
Create streams
|
Kernel with Streams
|
Kernel without streams
|
32x32
|
0.014075
|
0.017313
|
0.284529
|
0.262266
|
64x64
|
0.014052
|
0.017732
|
0.315197
|
0.297716
|
128x128
|
0.014728
|
0.018648
|
0.308442
|
0.400251
|
Time for different methods of evaluating neighbours
When we call a device function from within a thread we are not actually creating a new parallel block and thread to handle the process we are simply just calling a function in the same manner we could from the host. However when we use a kernel to handle the evaluation we need to launch a block and thread to handle the process, when using streams to launch the kernels concurrently we also have an extra overhead of having to create the streams.
Looking at the graph above we can see that we can call the device function to evaluate all 8 of our nodes neighbours quicker than we can create 8 streams to handle the neighbours. On top of the stream creating we also have to launch 8 kernels creating 8 blocks and threads to process the neighbors, this process in itself is extremely slow taking almost 18X longer than it does to call our device function.
Interestingly however it appears that using streams is potentially faster than not using streams, however this would really need to be tested for more graph sizes to get a fair results, this was not possible at the time due to errors in the timing code for large graph sizes.
The next change that was attempted involved each node storing a list of its neighbours in global memory on the device. In doing this it meant that it would be possible to call a single kernel which could be used to evaluate all of the 8 neighbours at the same time rather than calling an individual kernel for each neighbour as is needed when storing the nodes neighbours in local memory.
The next change that was attempted involved each node storing a list of its neighbours in global memory on the device. In doing this it meant that it would be possible to call a single kernel which could be used to evaluate all of the 8 neighbours at the same time rather than calling an individual kernel for each neighbour as is needed when storing the nodes neighbours in local memory.



No comments:
Post a Comment